You see, behavior is “hard wired”into our nervous system during development. The neuronal body plan is already present in our genome - the developmental genes that direct cells to grow into a specific type or in a specific direction. These genes during development take cues from the environment and hard wire innate behavior in to a species. I have always thought that to be the coolest thing. Think about it - we are not only a function of our genes but also product of our environment. And Dickson's research links the two together - how a presence of a single gene product directs the function of neurons responsible for sexual orientation in fruit fly males. Courtship behaviour in D. melanogaster is invovles a series of well chorographed steps that invovle the visual, the olfactory, the tactile, the acoustic, the gustatory and the mechanosensory stimuli being exchanged between the sexes (See Fig.1). The role of the female is more simplified -she simply runs away, gives the odd kick, then mates (or not).

Fig. 1 Courtship behaviour in Fruitfly
This normal courtship behaviour seems to be disrupted in the Fruitess mutants. Before we talk about what happens when we mutate this gene, lets us take a brief overview of what is known about the gene. Fruitless gene was molecularly cloned in 1996 and the putative protein encodes a transcription factor. Fruitless is sex specifically spliced - in lay man terms it means that males produce one version of this protein where as the females produce another. This sex specific splicing is regulated by presence or absence of another protein called Transformer, which in Drosophila also determines the sex in the fly.
So what happens if you produce the wrong version of the protein in either sexes? By forcing males to express the female-specific Fruitless transcript by using the awesome power of Fly genetics (:P), the Dickson Lab produced males that were sterile, uninterested in courting females, actively courting males, actually ending up forming courtship chains (see this). By contrast, females making the male version of the protein mated poorly, produced very few eggs, but — astonishingly — courted other females (see this), even to the point of forming chains.
So why does this happen? When you look at the the central nervous system of males and females ,there are very few differences in terms of sex-specific Fruitless expression- in number, position or wiring of cells that express this protein. But Fruitless is present in the olfactory sensory neurons which play an important role in fly courtship behavior. So when male fruit flies cannot produce this male specific form or produce a mutant form of this protein, you get males that court other males. In other words, a single gene encoded product is enought to shift the functioning of the nervous system from male to female mode, irrespective of the morphological sex of the animal. Simply put with mutant (rather non-sex specific)versions of the protein, flies change their sexual orientation but they not other aspects of their morphology.
Now the same gene is making a news splash again – the Kravitz lab has linked Fruitless to yet another sex-specific behavior – aggression/ fighting patterns. Aggression found in almost all animals - from sea anemones to human - helps to acquire food/shelter/ mates or defend the same. Despite its importance, relatively little is known of the neural and humoral mechanisms that are its proximate causes. Many behavioral patterns in aggressive behavior are shared in flies but there are a subset that are sex specific. Female fighting, for example, largely involves head butts and some shoving. Males show extended wing threats, wing-flicking while retreating, and high intensity components of fighting like boxing, tussling and holding. In contrast to male fighting behavior, no clear hierarchical relationship results from the interactions between female flies.
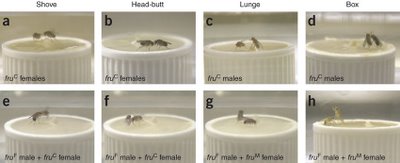 Figure 2. Aggresion Behavior in flies
Figure 2. Aggresion Behavior in fliesWhen the versions of Fruitless are swapped, the males fight like females (the sissies) and females lunge at their opponents as seen in the Figure 2 above. The top panel shows the normal aggresion patterns seen in males and females while the bottom panel shows what happens when the flies produce the wrong version of the protein. Panels e and f show males exhibiting female aggression pattern when they express the female version of the protein. When the sexes with the opppsite version of the protein are put together in the panel g and h. In panel g, the upright lunging fly is a female and so is the upright "boxing" fly in panel h , indicating that swapping the protien alters how the flies respond - another innate behaviour affected!
The question that still remains (the most important one) is what is the effector? What does Fruitless, a transcription factor, modulate in a gender specific manner to control the sex specific aspect of behaviour?
There is a lot to still uncover but we are finally beginning to glimpse at how a genes influence how we respond. I believe that most behaviour is hard wired but at the same time modulation of the behaviour is environmental dependant. And now we finally are beginning to tell the effect of nature on nurture. A fun time lies ahead in molecular neuroscience!
Reference
1 - Demir, E. & Dickson, B. J. Cell 125, 785−794 (2005).
2- Vrontou E, Nilsen, S. P., Demir, E., Kravitz, E. A. & Dickson, B. J. Nature Neuroscience - 9, 1469 - 1471 (2006)