The central dogma of molecular biology DNA-> RNA-> Protein shows the direction of flow of information of how the cells use the information stored in our DNA to make the necessary proteins. But the situation in most eukaryotes is a little more complex than that simple statement. In most eukaryotes, a gene sequence in a DNA is interrupted by non- coding information. Hence to make a protein, a cell first has to transcribe the gene (make a RNA copy of the gene, called pre-mRNA) and then modify the pre-mRNA by removing the non-coding sequence (intron) and joining the coding sequences (exons) together. The modified mRNA is then exported from the nucleus (where it was made) to the cytoplasm where the ribosome uses it as a template to make the protein. In simple English, the gene for making a proteinA looks like this "HEREabhjhdyfrhUSEndcbldfhdfmMEd ldshhglgmcFORdbfhdflhfnmc PROTEIN A". The task of the cells is to remove the gibberish and make a readable text out of the given instruction - HERE USE ME FOR PROTEINA. The cells then send this information to the ribosome (the protein factory) to make the protein.
Pre-mRNA splicing is the process in which the intronic sequences are removed within a large RNA-protein complex called spliceosome.
Why is splicing important? A spliceosme can remove the non-coding introns present in a given transcript varying combination in response to cellular cues, a process called alternative splicing. The recent completion of a draft of the human genome indicated that more than 59% of the human genes seem to be alternatively spliced (Hastings and Krainer,2001) and thus we can have more complexity (make a larger number of proteins) without increasing the number of genes present. For eg, the Dscam gene in flies has 38,000 alternatively spliced isoforms from four variable exon clusters!
More importantly, it is estimated that aberrant splicing causes about 15% of genetic diseases in humans (Philips and Cooper,2000). Thus, the spliceosome plays a critical role in generating the right template for making a protein and any abnormality in this process would be deleterious to the organism.
What do we know about this process? From genetic and biochemical experiments in the humble budding yeast, scientist have been able to understand how this process occurs. Because both the mechanism of splicing and the splicing machinery are highly conserved throughout eukaryotes, knowledge of yeast splicing gives us insights into the basic process in humans.
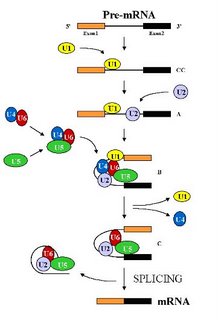 The spliceosome is the largest structure in the cell and is composed of five small nuclear RNAs ( called U1, U2, U4, U5 and U6 snRNAs) and over 100 different proteins (Stevens and Abelson , 2002). Under standard in vitro (i.e. in a test tube) assay conditions, the spliceosome assembles in a step wise manner through the addition of the U1 -> U2-> U4/U6.U5 snRNP particles (the small nuclear RNA along with its associated proteins, represented by a colored blob in the picture) on the pre-mRNA (See Figure). This assembly is an expensive process for the cell as each step consumes energy. But it also allows the apparatus to check each step and hence allows for a greater control over the overall process. Remember, a single mistake here would result in a protein that either does not function or functions abnormally. That to a cell would be hazardous and hence the cells err on the side of caution. After the assembly of the spliceosome, it undergoes structural rearrangements, resulting in the loss of U1 and U4 snRNAs, to become catalytically active (Brow D. A, 2002). Then, it proceeds to remove the intron by two transesterification reactions.
The spliceosome is the largest structure in the cell and is composed of five small nuclear RNAs ( called U1, U2, U4, U5 and U6 snRNAs) and over 100 different proteins (Stevens and Abelson , 2002). Under standard in vitro (i.e. in a test tube) assay conditions, the spliceosome assembles in a step wise manner through the addition of the U1 -> U2-> U4/U6.U5 snRNP particles (the small nuclear RNA along with its associated proteins, represented by a colored blob in the picture) on the pre-mRNA (See Figure). This assembly is an expensive process for the cell as each step consumes energy. But it also allows the apparatus to check each step and hence allows for a greater control over the overall process. Remember, a single mistake here would result in a protein that either does not function or functions abnormally. That to a cell would be hazardous and hence the cells err on the side of caution. After the assembly of the spliceosome, it undergoes structural rearrangements, resulting in the loss of U1 and U4 snRNAs, to become catalytically active (Brow D. A, 2002). Then, it proceeds to remove the intron by two transesterification reactions.
 The resultant message is released from the spliceosome along with the intron. The spliced RNA is exported to the cytoplasm for translation into the protein and the intron degraded by enzymes in the cell. The spliceosome is disassembled and the components (proteins and the snRNAs) recycled for another round of splicing.
The resultant message is released from the spliceosome along with the intron. The spliced RNA is exported to the cytoplasm for translation into the protein and the intron degraded by enzymes in the cell. The spliceosome is disassembled and the components (proteins and the snRNAs) recycled for another round of splicing.
Though much is known about the overall process, there is no insights into what triggers the activation. What informs the spliceosome that everything is set in place and hence go ahead and splice? How does the cell control the ATP driven helicases that remodel the spliceosome at each step? Or what cues the cell about abnormal spliceosome and how does it take a stalled spliceosome apart?
Next time I will try and address the role splicing plays in Humans. How does a cell choose which exon to keep? How do DNA elements present in the gene (ISEs) affect choice of exon? Does the rate at which the transcript is made affect exon choice? So keep your eyes out for Splicing -part deux.
References -
Brow D. A, Annu Rev Genet., 2002, Jun 11; 36:333-60.
Hastings and Krainer, Curr Opin Cell Biol., 2001, Jun; 13(3):302-9
Philips and Cooper, Cell Mol Life Sci., 2000, Feb;57(2):235-49
Stevens and Abelson , Methods Enzymol. 2002;351:200-20.
Check this Animation
9 comments:
Does the intron portion have any function or raison d'etre in an organism?
A very valid question. For a long time most of us thought that Introns were insignificant - I mean, if they are not useful to make the protein, they are basically useless. But slowly the introns are being vindicated. They play an enormous role in gene regulation.
I will touch upon this in a follow up post. The post become too long and as my Boss says, if you talk for more than 5 min - people are likely to forget the first 4. So I am breaking up this fascinating topic into a number of posts.
The Boss? He sounds like an underworld don.
@ Raindrop - Yeah, He is! I keep waiting for him to say "I will make you deal you cant refuse" in the Godfather voice..
Heck, Meetings are going to be fun from now on :)
Sakshi,a very complicated process written in a sinple manner. I am sure a non-bio can understand it now. But did remind me when I was studying mol-bio paper in MSc. haven't thought about u1, u4/u6 in a long while.
Hey write a post on RNAi so that when 'D' asks me Qs next month I am ready. :)
Very interesting post and very nicely written. I have a bunch of questions, but I will ask only 4 as we could go on forever:
1. How much of the picture you mentioned above is the general picture in eukaryotes and how much of it is yeast specific? That is how conserved are these RNA non-coding genes and other regulation units in nature?
2. Are there other kinds of regulation complexes (protein, RNA, protein-RNA) found in yeast and how different are they? How different is this picture from bacteria/archaea translation regulation?
3. Do U1 and U2 bind to sequence specific regions and how specific is this region? In other words, do introns in all genes start and end with the same sequence?
4. Silly question but why no U3?
@ BaL - Most of the core machinery is higly conserved. What you find in yeast will have a counterpart in Humans. They are few yeast specific proteins and since Humans undergo alternative splicing, they have many proteins that are not found in yeast. That being said, the core machinery is found almost protein for protein.
3. Good question. U1, U2 and U6 bind specific sequences on the mRNA. These are highly conserved. I will write more on this in the part 2. Just now suffice it to say that there are conserved elements on the pre-mRNA that are recognized by the snRNA.
3. They were named as they were discovered. There is a U3 but it has no role in this. In yeast it is invovled in snRNA regulation.
For ques no 3 - can you be more specific?
Just wondering whether there was alternative machinery other than just U1, U2, U4, U5 & U6 (either completely new machinery or another splicing complex that uses some of these RNA molecules and, in addition, need other RNA/proteins)..
@ Bal - yes there is. It is called the minor spliceosome. It uses the same U2 and U5 but the rest are called "atac" snRNAs. These splice a subset of the RNA which have modified splice sites.
Too lazy to sign in Sakshi
Post a Comment